Interesting News About Aging
Rethinking estrogen supplementation: anti-aging effects of postmenopausal estrogens
Especially since the report of the so-called Women’s Health Initiative (WHI)—a large randomized clinical trial of estrogen and progestin supplementation - was published 23 years ago, peri- and postmenopausal estrogen replacement therapy has remained a somewhat controversial subject. Now, however, Eric Topol, MD, in his Ground Truth podcast and Substack newsletter—a steady and very reliable source of the VOICE—in detail described interesting new data suggesting that postmenopausal estrogen supplementation may, indeed, have anti-aging effects and, therefore, may require a readjustment in our thinking about this treatment. More specifically, the evidence came from organ clocks, as we have already on repeated occasions reported in The Reproductive Times, an increasingly popular study tool in anti-aging medicine.1
The WHI was stopped prematurely because supplementation in an interim analysis demonstrated a significant risk of invasive breast cancer (see the front-page article in The New York Times at the time). But the results were more complex: They suggested an increased risk for coronary heart disease, stroke, and pulmonary embolism, in addition to the above-noted invasive breast cancer risk, and reduced risks for colon cancer and hip fractures. As Topol noted, though statistically significant, the absolute risks were really quite small (19 per 10,000 person-years). While the relative increase in risk for breast cancer was 26%, the absolute risk was only 8 cases per 10,000 women-years.
The consequences of the WIH report were, nevertheless, substantial: Postmenopausal hormone supplementation—based on prescription frequency—declined by over 60%.
Without going into further detail (for that, we refer our readers to Topol’s article), the interpretation of the WHI data significantly changed over time, recognizing significant earlier shortcomings. Among those was the observation that supplementation initiated before age 60 or within 10 years of menopause actually reduced risks of heart disease and all-cause mortality. Moreover, observational data (i.e., lower levels of evidence) also supported benefits on cognitive health, lower Alzheimer’s disease risk, and Type 2 diabetes. Combined, these findings could be interpreted as a beneficial anti-aging effect of estrogen.
Topol then addressed several studies in the literature and noted that among 9 drugs found to be associated with reduced all-cause mortality, 4 were estrogen preparations. And a very recent study of 460,000 women with a mean age of 42 years revealed that estrogen supplementation (alone) was linked to reduced young-onset breast cancer.
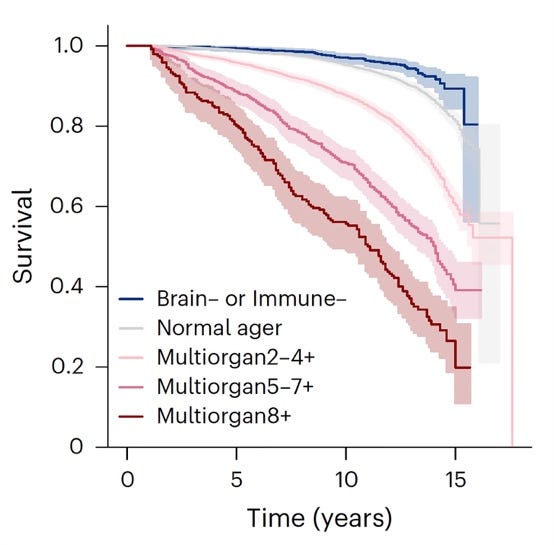
The apparent incentive for Topol’s article was, however, a preprint at bioRxiv 2 scheduled to appear practically unchanged in Nature Medicine in the journal’s July 9 issue, which reported the following rather remarkable findings: It has been established that organ-derived plasma protein signatures derived from protein arrays can track organ-specific aging, disease, and mortality in humans. How robust these associations are, to what degree these models can offer clinical utility, and their biological underpinnings, however, remain largely unknown. The group of international investigators of this study, therefore, estimated the biological age of 11 organs from 44,526 individuals in the UK Biobank using an antibody-based proteomics platform to model disease and mortality risk using ~3,000 plasma proteins in study participants followed for approximately 17 years.
They found the following: Organ age estimates were “organismal” (not organ-specific), associated with future onset of heart failure, chronic obstructive pulmonary disease, type II diabetes, and Alzheimer’s disease, and sensitive to lifestyle factors such as smoking and exercise, hormone replacement therapy, or supplements.
Remarkably, the accrual of aged organs progressively increases mortality risk, while a youthful brain and immune system are uniquely associated with disease-free longevity (see figures above). These findings support the use of plasma proteins for monitoring organ health and the efficacy of drugs targeting organ aging disease, and found the brain and immune system clocks to be most influential.
Topol concluded that the protective effect of estrogen largely accounted for why women develop heart disease on average 10 years after men. From mouse experiments, we also have learned that health span can likely be prolonged by delaying physiological ovarian failure (i.e., menopause). And quoting another author, Topol noted that the ovary is the only organ in the human body expected to fail prematurely in comparison to all other organs. At least in mouse models life span of an old mouse that received a young ovary transplant is extended. Topol, therefore, suggests that estrogen supplementation may, after all, be “an attractive alternative.”
References
Topol E. Ground Truths. July 5, 2025.
Se-Hwee et al., bioRxiv. 2025. https://doi.org/10.1101/2024.06.07.597771
An interesting update on the epigenetic aging of women during pregnancy
That pregnancy epigenetically ages women has been known for some time. Now a recent paper by Stanford University investigators in Obstetrics & Gynecology added some interesting detail: In a prospective cohort study of nulliparous women (age 18-50 years) seeking obstetric (pregnant 10-14 weeks) or gynecologic (nonpregnant) care in 2020-2021, they collected blood samples at enrollment (time 1) and postpartum day 1 (pregnant, time 2) or 7 months later (nonpregnant, time 2). Epigenetic age was measured with 11 established clocks from Illumina EPIC 2 arrays. Results were then scaled per 200-day interval. And P values were adjusted for multiple testing.
And then, as the most interesting part of the study, the investigators, through multivariable logistic regression explored associations between first-trimester epigenetic age and a composite of potentially immune-mediated complications (hypertensive disorders, gestational diabetes mellitus, preterm birth before 37 weeks of gestation, and small-for-gestational-age birth weight) adjusted for age and body mass index (BMI) higher than 30 at time 1.
And these were the results: The study enrolled 75 women, 45 (60.0%) pregnant. A total of 61 (81.3%) completed the study. Compared with nonpregnant women, pregnant women exhibited significant epigenetic age acceleration in six clocks (Hannum, PhenoAge, GrimAge, GrimAge2, Stem Cell Division, DunedinPACE). Moreover, epigenetic age acceleration per 200 days in the pregnant cohort ranged from 1.58 years (Hannum, 95% CI, 0.45-2.72, P=.01) to 5.28 years (PhenoAge, 95% CI, 2.97-7.61, P<.01). Each additional year of first-trimester GrimAge2 increased odds of the composite of pregnancy complications by 36% (adjusted odds ratio [aOR] 1.36, 95% CI, 1.01-1.84), while chronologic age (in continuous years) showing no association (aOR 1.00, 95% CI, 0.83-1.21).
Though not earthshaking news, these data are interesting and can be summarized in the following way: Within-person epigenetic aging was accelerated by pregnancy by up to 5.3 years. Older first-trimester GrimAge2 (an epigenetic biomarker of human mortality and morbidity risk), but not chronologic age, was associated with a composite of several pregnancy complications. These findings obviously encourage further investigations along the lines explored by this paper. At the same time, the data, however, also suggest caution about possible overinterpretations: Obviously, the various epigenetic clocks demonstrated different severities of the pregnancy impact. For obstetrical care, however, even more importantly, the study found outcome significance only in a composite of pregnancy complications and not for individual pregnancy complications.
This finding has two possible explanations: either the impact of individual complications is very limited, or the power of the study was too low to show significance for individual pregnancy complications. Which of these two potential explanations applies is, of course, important to determine.
Reference
Panelli et al., Obstet Gynecol. 2025; PMID: 40638919; PMCID: PMC12252219 (available on 2025-07-10). DOI: 10.1097/AOG.0000000000006000. Online ahead of print.
The latest in BASIC SCIENCES
Interesting news regarding DNA double-stranded break repair
Double-stranded break repair is, of course, a very essential biological process, with considerable physiological consequences if it malfunctions, as we elsewhere in the genetics section of this literature review—for example—briefly noted in women with BRCA1 and BRCA2 mutations who, as a consequence, demonstrate lower than normal functional ovarian reserve.
A recent paper in Molecular Cell by British investigators1 started with the statement that the DNA’s double-stranded breaks are extremely toxic genomic lesions, which human cells have learned over evolution to repair. Such repairs happen because double-stranded breaks induce histone modifications (and a recruitment cascade of repair proteins regulated by histones and repair protein modifications of phosphorylation, acetylation, methylation, ubiquination), and—now coming to the main subject of the paper—the conjugation of small ubiquitin-like modifiers called SUMO1-3 in a process called SUMOylation.
As the paper also notes, amplitudes of small-modifier protein signaling through ubiquitin and small ubiquitin-like modifiers (SUMO1-3) are critical for the correct phasing of DNA repair protein that accumulates, their activity, and clearance, and—ultimately—for completion of DNA double-strand break repair. In this paper, the investigators now demonstrated that the—so far only poorly explored SUMO isoform, SUMO4—enhances the deSUMOylation activity of the SUMO protease SENP1, thereby restraining excessive cellular SUMOylation. This then prevents excessive recruitment of the SUMO-binding protein RAP80 to damage sites and promotes DNA repair. In short: (i) SUMO4 conjugation is deleterious; (ii) SUMO4 promotes SENP1 catalytic activity; and (iii) SUMO4:SeNP1 together restricts SUMO signaling and accumulation of the BRCA1-A complex.
And here is another recent paper from the same British group of investigators, but this time, in Nature Communications (not a bad record!).2 Unsurprisingly, the group adds further information regarding double-strand break repair response. This time, the data relate to RNF168, an E3 ubiquitin ligase critical for the repair response. This protein is recruited to ubiquitin and amplifies ubiquitin signals at damaged chromatin. If not properly regulated, it can initiate an uncontrolled ubiquitin cascade, which would hurt the repair process.
Without going into too much detail, the authors here defined a mechanism of direct RNF168 regulation that is part of the normal damage response, promoting RNF168 regulation dissociation from chromatin and limiting deleterious ubiquitin signaling while promoting correct ubiquitin signaling and DNA repair.
References
Garvin et al., Molecular Cell 2025;85:877-8
Chauhan et al., Nat Commun 202516:3399
Are we rewriting the rules of cell division?
This is at least what a commentary in Science argues1 in relevance to a paper out of Nicholas Plachta, PhD’s laboratory, which recently appeared in the same issue of Science, suggesting that at preimplantation stages of embryos, actin organizes chromosomes and microtubules to ensure mitotic fidelity.2
The authors of the paper noted that at preimplantation stages, a single copy of the genome is going through several rounds of mitotic cell division. Errors during mitosis can result in mis-segregation of chromosomes and aneuploidy of cells, which means abnormal chromosome numbers. What maintains chromosomal fidelity, therefore, has remained a core issue in reproductive biology.
The traditional opinion has been that the spindle apparatus manages chromosome organization. More specifically, the believe has been that microtubules in principle organize the process of mitosis. More recently, this—what the authors call the microtubule-centric view—has, however, been increasingly challenged by the recognition that cytoplasmic actin filaments may also play an important role. Using high-resolution imaging, the authors have now revealed in their paper that an actin cytoskeleton actually organizes chromosomes and microtubules during early embryonic development. They identified two actin assemblies that performed functions usually attributed to the centromere. They also noted that this finding finally explains how mouse embryos “can do it”—without having the machinery for the longest time assumed to be alone responsible for mitosis. Truly, never a dull moment in reproductive biology!
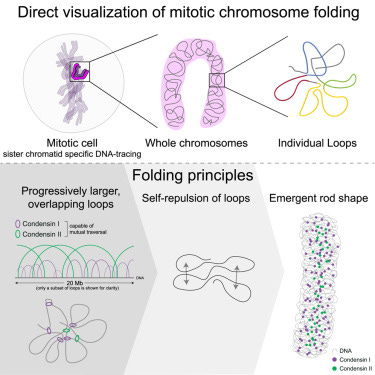
And if we are already talking about mitosis, here is a paper that traced nanoscale DNA only to reveal the self-organization mechanism of mitotic chromosomes.3 What they found they summarized as follows: 3D DNA tracing resolved the internal architecture of mitotic chromosomes. Chromosomal rod shape then emerged without a contiguous scaffold or regular handed helix, and condensin transversal enabled extrusion of overlapping loops of up to several megabases. Finally, self-repulsion of overlapping loops generates a mitotic chromosomal rod shape.
How genomic DNA folds during cell division to form the characteristic rod-shaped mitotic chromosomes essential for faithful genome inheritance has remained an open question. The authors’ structural analysis revealed a characteristic genome scaling minimum of 6–8 megabases in mitosis. Data-driven modeling and molecular perturbations then helped them to demonstrate that—formed by condensins—very large and strongly overlapping loops are the fundamental structure of mitotic chromosomes. These loops compact chromosomes to the limit set by chromatin self-repulsion. Their characteristic length, density, and increasingly overlapping structure observed in 3D fully explained how a rod-shaped mitotic chromosome structure then emerges by self-organization during cell division (see Figure).
References
1. Bement M. Science 2025;388(6749), 817-818
2. Hernandez et al., Science 2025;388(6749)835
3. Sandvold Beckwith et al., Cell Press 2025;188(10):P2656-2669
Always something new to learn in Drosophila melanogaster
A Preprint paper in eLife also attracted our attention because it identified a female germline-specific protein that is crucial for germline stem cell renewal and differentiation, as well as oogenesis.1 Specifically, this “valuable” study in the eLife assessment reports the first characterization of the CG14545 gene in Drosophila melanogaster, in the process naming the gene “Sakura.”
The gene appears to be essential for oogenesis as well as female fertility in general and is mostly expressed in ovaries, and there in germline cells, inclusive of germline stem cells. Female null mutants of the gene display only rudimentary ovaries without germlines and tumorous phenotypes. They are unable to produce eggs and, of course, are fully sterile. Finally, the authors also identified Out (Ovarian Tumor) as a protein binding partner of Sakura, defining both seemingly as partners in germline stem cell renewal and differentiation, as well as oogenesis
Since the Drosophila female germline is an excellent model for the study of regulatory mechanisms in the germline, these findings may have considerable significance for the human experience as well.
Reference
Azlan et al., Reviewed and preprint eLife. https://doi.org/10.7554/eLife.103828.3